R Interface to HDF5
HDF5 1.8.14 has been pre-compiled for Windows and is available at - thus no manual installation is required. The latest release version of hdf5r can be installed from any CRAN Mirror using the R command. Install.packages('hdf5r') For the latest development version from Github you can use. Headers are 1.10.4, library is 1.10.5 SUMMARY OF THE HDF5 CONFIGURATION General Information: - HDF5 Version: 1.10.5 Configured on: 2019-03-04 Configured by: Visual Studio 14 2015 Win64 Host system: Windows-10.0.17763 Uname information: Windows Byte sex: little-endian Installation point: C:/Program Files/HDF5 Compiling Options: - Build Mode: Debugging Symbols: Asserts: Profiling: Optimization Level: Linking Options: - Libraries: Statically Linked Executables: OFF LDFLAGS. INSTALL - Start Here. General instructions for compiling and installing the library INSTALLCMAKE - instructions for building with CMake (Kitware.com) INSTALLparallel - instructions for building and configuring Parallel HDF5 INSTALLWindows and INSTALLCygwin - MS Windows installations.
Bioconductor version: Release (3.12)
This package provides an interface between HDF5 and R. HDF5's main features are the ability to store and access very large and/or complex datasets and a wide variety of metadata on mass storage (disk) through a completely portable file format. The rhdf5 package is thus suited for the exchange of large and/or complex datasets between R and other software package, and for letting R applications work on datasets that are larger than the available RAM.
Author: Bernd Fischer [aut], Mike Smith [aut, cre] , Gregoire Pau [aut], Martin Morgan [ctb], Daniel van Twisk [ctb]

Maintainer: Mike Smith <mike.smith at embl.de>
Install Hdf5 Library Linux
Citation (from within R, enter citation('rhdf5')):
Installation
To install this package, start R (version '4.0') and enter:
For older versions of R, please refer to the appropriate Bioconductor release.
Documentation
To view documentation for the version of this package installed in your system, start R and enter:
| HTML | R Script | Reading HDF5 Files In The Cloud |
| HTML | R Script | rhdf5 - HDF5 interface for R |
| HTML | R Script | rhdf5 Practical Tips |
| Reference Manual | ||
| Text | NEWS |
Details
| biocViews | DataImport, Infrastructure, Software |
| Version | 2.34.0 |
| In Bioconductor since | BioC 1.6 (R-2.1) or earlier (> 16 years) |
| License | Artistic-2.0 |
| Depends | R (>= 3.5.0), methods |
| Imports | Rhdf5lib(>= 1.11.0), rhdf5filters |
| LinkingTo | Rhdf5lib |
| Suggests | bit64, BiocStyle, knitr, rmarkdown, testthat, microbenchmark, dplyr, ggplot2 |
| SystemRequirements | GNU make |
| Enhances | |
| URL | https://github.com/grimbough/rhdf5 |
| BugReports | https://github.com/grimbough/rhdf5/issues |
| Depends On Me | GenoGAM, GSCA, HDF5Array, HiCBricks, LoomExperiment |
| Imports Me | BayesSpace, BgeeCall, biomformat, bnbc, bsseq, CiteFuse, cmapR, CoGAPS, CopyNumberPlots, CRISPRseek, cTRAP, diffHic, DmelSGI, DropletUtils, EventPointer, FRASER, GenomicScores, gep2pep, h5vc, HiCcompare, hicrep, IONiseR, MafH5.gnomAD.r3.0.GRCh38, MethylSeqData, MOFA, MOFA2, phantasus, PureCN, recountmethylation, ribor, scCB2, scone, signatureSearch, signatureSearchData, slinky |
| Suggests Me | edgeR, slalom, Spectra, SummarizedExperiment, tximport, zellkonverter |
| Links To Me | |
| Build Report |
Package Archives

Follow Installation instructions to use this package in your R session.
| Source Package | rhdf5_2.34.0.tar.gz |
| Windows Binary | rhdf5_2.34.0.zip (32- & 64-bit) |
| macOS 10.13 (High Sierra) | rhdf5_2.34.0.tgz |
| Source Repository | git clone https://git.bioconductor.org/packages/rhdf5 |
| Source Repository (Developer Access) | git clone git@git.bioconductor.org:packages/rhdf5 |
| Package Short Url | https://bioconductor.org/packages/rhdf5/ |
| Package Downloads Report | Download Stats |
This is just a quick walkthrough of installing HDF5 on a linux system. I'm doing this on CentOS 6.5 right now, but I'm pretty sure the steps were the same for Ubuntu. There's an RPM package for RedHat/CentOS, but since the tgz should work on more systems, that's what I'm using here.
First, download the latest appropriate .tgz package for your system from http://www.hdfgroup.org/HDF5/release/obtain5.html – in this case it's http://www.hdfgroup.org/ftp/HDF5/current/bin/linux-x86_64/hdf5-1.8.13-linux-x86_64-shared.tar.gz
Install The Hdf5 Library Version
Make a place in the filesystem for the hdf5 tools to live:

Extract the tools:
Make a symbolic link “latest” to point to the latest (only, at this point) version installed:
Run the h5redeploy script to change the paths in a few scripts:
Add /opt/hdf5/latest/bin to the $PATH for connections after this point:
Install Hdf5 Library Python
Log out and back in, then check your path:
[lariosadd@hydra ~]$ echo $PATH
/usr/lib64/qt-3.3/bin:/usr/local/bin:/usr/bin:/bin:/usr/local/sbin:/usr/sbin:/sbin:/opt/cuda/latest/bin:/home/lariosadd/bin:/opt/cuda/latest/bin:/opt/hdf5/latest/bin
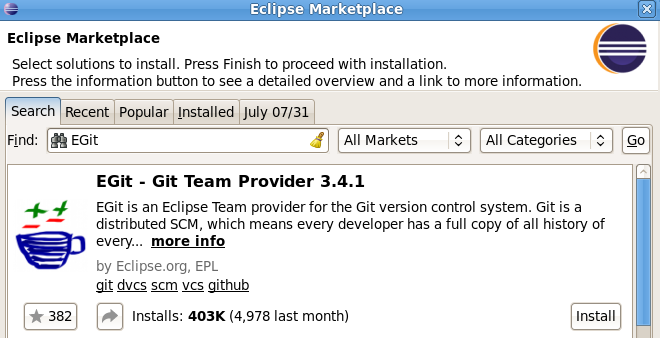
Looks good, so see if it works, I guess?
Comments are closed.